Topology prediction algorithms
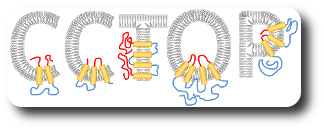
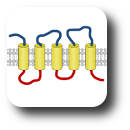 The group leader has been engaged in sequence analysis of transmembrane proteins since the end of the 1990s and achieved remarkable results in the field of topology prediction. In a recent research, we developed an algorithm that incorporates the prediction results of the most accurate state-of-the-art prediction methods as well as experimentally established topology data and other computational biology results. The developed constrained, consensus prediction method (CCTOP) is the most accurate prediction method so far, therefore it was used for creating the human transmembrane proteome (HTP database).
The group leader has been engaged in sequence analysis of transmembrane proteins since the end of the 1990s and achieved remarkable results in the field of topology prediction. In a recent research, we developed an algorithm that incorporates the prediction results of the most accurate state-of-the-art prediction methods as well as experimentally established topology data and other computational biology results. The developed constrained, consensus prediction method (CCTOP) is the most accurate prediction method so far, therefore it was used for creating the human transmembrane proteome (HTP database).
People working on this project
| Gábor E. Tusnády |
| László Dobson |
Related publications
|
1.
Dobson, L, Szekeres, LI, Gerdán, C, Langó, T, Zeke, A and Tusnády, GE
(2023)
TmAlphaFold database: membrane localization and evaluation of AlphaFold2 predicted alpha-helical transmembrane protein structures.
Nucleic Acids Res
51:
D517-D522.
|
|
2.
Dobson, L, Reményi, I and Tusnády, GE
(2015)
The human transmembrane proteome.
Biol Direct
10:
31.
|
|
3.
Dobson, L, Reményi, I and Tusnády, GE
(2015)
CCTOP: a Consensus Constrained TOPology prediction web server.
Nucleic Acids Res
43:
W408-12.
|
|
4.
Tusnády, GE and Simon, I
(2001)
The HMMTOP transmembrane topology prediction server.
Bioinformatics
17:
849-50.
|
|
5.
Tusnády, GE and Simon, I
(1998)
Principles governing amino acid composition of integral membrane proteins: application to topology prediction.
J Mol Biol
283:
489-506.
|
|
6.
Dobson, L, Gerdán, C, Tusnády, S, Szekeres, LI, Kuffa, KR, Langó, T, Zeke, A and Tusnády, GE
(2024)
UniTmp: unified resources for transmembrane proteins.
Nucleic Acids Res
52:
D572-D578.
|
Recent publications
News
| 2020-06-02 - Paper published |
| 2019-03-08 - PhD degree |
| 2019-02-07 - Paper published |
| 2018-11-14 - Scientific Students' Association third prize |
| 2018-10-30 - Paper published |
